A decades-old reaction that has become the poster boy for the field of 'click chemistry' is now expanding into biology, as Hayley Birch discovers
A decades-old reaction that has become the poster boy for the field of ’click chemistry’ is now expanding into biology, as Hayley Birch discovers
Speaking in December 1960 at the Royal Institution of London, the German chemist Rolf Huisgen explained how his work on one particular type of reaction had at first proceeded at a ’snail’s pace’, employing just two people. But by the third year, he went on to boast, his team had expanded to 17. How gratified he must feel today to know that ’Huisgen’s reaction’ - otherwise known as 1,3-dipolar cycloaddition - is the focus of a burgeoning field of chemistry and that his work has been cited in many thousands of scientific papers.

Huisgen’s reaction has come to exemplify the type of powerful, simple to perform, and highly reliable reaction that embodies ’click chemistry’ - a term invented in 2001 by Barry Sharpless from the Scripps Research Institute in La Jolla, US. React an alkyne with an azide in the presence of copper and - click! - they snap together like Lego bricks. Huisgen’s reaction, like other click chemistry reactions, benefits from favourable thermodynamics that drive it forwards under mild conditions to produce high yields of a single product. Sharpless’ original motivation for promoting click chemistry was to encourage development of simpler strategies for medicinal chemists to use in drug synthesis,1 but, nearly a decade on, it is biologists who are adopting click chemistry reactions - and in particular Huisgen’s.
Great gel
At the Howard Hughes Medical Institute in Colorado, US, Cole DeForest has been employing cycloaddition reactions to make gels for culturing cells in 3D that he hopes will one day form the basis of a tissue regeneration platform. Click chemistry provides a more direct route to creating the gel.
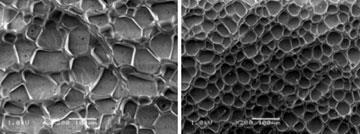
DeForest’s lab has been making hydrogels for some 15-20 years. ’A lot of the work in our lab has been dealing with trying to encapsulate different types of stem cells and trying to have the material induce differentiation of these cells, ultimately for implantation in human patients,’ he explains. Hydrogels, says DeForest, can be designed to dictate the sort of behaviour that the cells elicit - how they spread and adhere to surfaces, and, crucially, how they differentiate. All sorts of components can be added to the hydrogel to influence these behaviours. With the hydrogel as the base material, alkyne and azide groups act like Velcro patches for sticking on the various molecules that the team wants to test out.
Huisgen’s original scheme, however, hasn’t remained entirely intact - the simple alkyne-azide reaction favoured by chemists is not fully compatible with biological systems. As Renier van der Hoorn, a plant biologist at the Max Planck Institute for Plant Breeding Research in Cologne, Germany, points out, Huisgen’s original reaction wasn’t meant for studies in live cells. ’You need copper for the traditional click chemistry and copper denatures proteins and is simply toxic,’ he says.
Probing plant enzymes
Van der Hoorn’s own team has recently focused on using small molecule probes to label plant enzymes, for instance, showing the activity of the enzymes in leaves. Click chemistry avoids using larger fluorescent labels that are incapable of permeating cell membranes. Instead, a small alkyne or azide modified tag is added in vivo before making an extract and then adding a label, which is designed to ’click’ with the tag using Huisgen’s old trick of reacting azide with alkyne. But because the copper catalyst is toxic to plants, the reaction can not be done in live tissue. And as van der Hoorn notes, copper also denatures their purification columns.
Unlike study of biochemical processes, tissue regeneration requires live cells and DeForest’s team was looking for a method that would be compatible with their differentiating stem cells.
Copper-free clicks
In 2007, a paper by Jeremy Baskin at the University of California, Berkeley, US, and colleagues presented a solution to the problem - copper-free click chemistry performed in live cells.2 Based on the classic Huisgen cycloaddition, Baskin’s revamped version placed the alkyne group in a ’strained’ cyclooctyne ring, adding fluorine atoms to withdraw electrons. The idea was to put tension on the click components to make them more reactive and to drive the reaction without the need for a catalyst.
In 2008, Baskin and colleagues went on to prove that the method worked in live animals when they used it to watch cells and tissues forming in zebrafish embryos (Chemistry World, June 2008, p31). They achieved this by snapping fluorescent tags onto sugars in the cell membranes of the fish - the sugars acted as indicators of developmental processes.
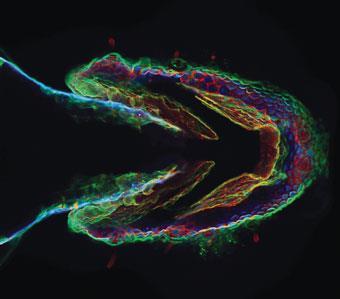
The ring strain method was soon adopted by DeForest’s lab. Last year, DeForest was lead author of a study published in Nature Materials describing a method for creating hydrogels using copper-free click chemistry.3 ’We end-functionalise polymers and synthetic peptides with these chemical moieties and then we mix the polymers and the peptides together and they undergo this click reaction which is responsible for forming the hydrogels,’ says DeForest. ’And because we’ve done this with a copper-free click chemistry, we can actually form these hydrogels in the presence of cells without sacrificing viability.’ The advantage over traditional methods, he says, is that the click reaction is high yielding, quick and doesn’t form any by-product - if it did, this could also potentially be toxic to cells.
Patterning hydrogels
But that’s not all. A second click reaction is used to ’pattern’ biological functionality into the 3D network. Straying away from the gold standard Huisgen’s reaction, the team employed a light-induced thiol-ene reaction - the addition of a thiol to an alkene - allowing them to snap any thiol-containing compound of their choice onto the main body of the hydrogel. Thus, by incorporating a thiol into an adhesive molecule and shining a light on specific areas, they were able to pattern in attachment sites for cells and show that the cells did, indeed, attach to them. Few technologies have this level of control, claims DeForest.
The team is now working on being able to pattern in more exotic molecules such as growth factors and peptides that may induce differentiation. ’We envision that we will be able to encapsulate a uniform population of stem cells into our hydrogel and then pattern in different growth factors to induce different lineages,’ says DeForest. ’So we will be able to create very complex tissue-like structures of spatially controlled, differentiated stem cells.’
Tracking devices
While DeForest attempts to generate tissues from scratch, click chemistry methods are being developed by neurobiologists to track dividing cells in naturally regenerating brain tissue. Adrian Meedeniya, a neurobiologist at Griffith University in Brisbane, Australia, has based his work on that of a team at Harvard Medical School, which published a method for detecting DNA synthesis using click chemistry in 2008.4 The method involved creating an analogue of one of the nucleosides contained in DNA - uridine - with an all-important alkyne group, which was exposed in the groove of the DNA helix. The researchers ’clicked’ this with a fluorescent azide that could be imaged to track DNA synthesis and therefore cell division.

Meedeniya considers this work to be ’seminal’. He has since employed 5-ethynyl-2 ? -deoxyuridine (Edu), a uridine analogue, to track cell division in embryonic mouse brains. So far though, his methods aren’t completely compatible with live cell systems; the next step, he says, will be removing copper from the equation. But Edu does improve significantly on its predecessor Brdu, which is detected by a fluorescent antibody. As Meedeniya explains, to detect this antibody the DNA has to be unzipped, requiring a fairly harsh protocol and resulting in complete destruction of the cell’s molecular profile. In other words, genetic and proteomic information about the cell that could be important later is lost. ’That was our major hurdle, and with Edu and the click chemistry protocol we can conserve the molecular profile,’ says Meedeniya. ’We don’t have to enable it too much or even unzip the DNA. It’s a very quick, easy protocol and a really powerful tool for looking at dividing cells.’
So powerful that Meedinya is working on a series of studies using Edu to try to unravel aspects of various diseases. Some focus on neurodegenerative disorders, such as Parkinson’s, while one is aimed at revealing the mechanisms that regulate cell division in glioblastoma multiforme, the most aggressive form of brain cancer. He is also helping to develop click chemistry products that will function as standard assays and may be invaluable in stem cell and cancer research. (Several Click-it products are already marketed by California, US-based biotechnology company, Life Technologies.)
Room for more
But are too many chemists and biologists still clinging to Huisgen’s reaction? It is, after all, celebrating its 50th birthday this year. ’I think there are certainly many other [click] reactions,’ says DeForest. ’There’s definitely room for pursuing more biologically compatible click reactions, that’s for sure.’ In theory, he adds, click reactions are supposed to be orthogonal to one other, meaning one reaction can be performed in the presence of other functionalities without undergoing side reactions. So discovering more click reactions could mean gaining more control over a system but also providing scope for multiple staining, allowing researchers to track different populations of cells at the same time.
As for Sharpless’ original intention of developing easy-to-make drugs, several companies are already rumoured to be working on click products, including Intezyne in Florida. The company is using click methods to modify cancer drugs in such a way that they can be better targeted at tumours. Meanwhile, a group at the National University of Singapore led by Shao Yao has adopted click chemistry techniques to rapidly synthesise inhibitors for enzymes thought to be involved in chronic myelogenous leukemia and to help identify targets for malaria drugs (see box, p38). The growing compatibility of click chemistry with biology suggests its potential is greater than Sharpless, and certainly Huisgen, would ever have guessed.
Leukaemia drugs
Click chemistry can be used to rapidly assemble a library of hundreds of potential drug candidates. Last year, Shao Yao and colleagues at the National University of Singapore did just that to generate candidates for the treatment of chronic myelogenous leukaemia (CML)5 - which affects the white blood cells and is responsible for up to a fifth of all cases of leukaemia. The only known cure is a bone marrow or stem cell transplant.
Several drugs have, however, been approved that can prolong survival, and Yao’s team was able to identify a few more potentials from its screens. The approach was based on targeting Abelson tyrosine kinase, an enzyme implicated in CML, and using Huisgen’s classic alkyne-azide reaction.
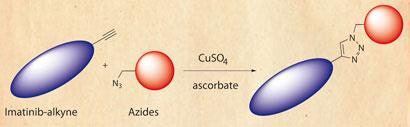
First, as proof of principle, the researchers produced a library of molecules through click reactions between an adenosine diphosphate (ADP)-alkyne and 344 different azides. Then they took imatinib, the first-line therapy for CML patients, and swapped its piperazine moiety for an alkyne, generating a clickable molecule with an already proven drug attached. This was then snapped onto 45 different azides and the products ability to inhibit Abelson tyrosine kinase tested, with some showing a slight improvement over the original imatinib.
According to Yao, his team is already making second and third generations of its candidate drugs. ’Our new generations focus on improving the binding property of the probes, for instance, by using more potent drug templates such as dasatinib instead of imitinib,’ he says.
Hayley Birch is a science writer based in Bristol, UK
References
1 H C Kolb et al, Angew. Chem. Int. Ed., 2001, 40, 2004
2 J M Baskin et al, Proc. Natl. Acad. Sci. USA, 2007, 104, 16 793 (DOI: 10.1073/pnas.0707090104)
3 C A DeForestet al, Nature Mater., 2009, 8, 659 (DOI: 10.1038/nmat2473)
4 A Salic and T J Mitchison, Proc. Natl. Acad. Sci. USA, 2008, 105, 2415 (DOI: 10.1073/pnas.0712168105)
5 K A Kalesh et al, Org. Biomol. Chem., 2009, 7, 5129 (DOI: 10.1039/b913333j)






No comments yet