Ring-shaped extrachromosomal DNA is implicated in many cancers. Rachel Brazil talks to the scientists trying to uncover their secrets
-
Discovery and prevalence: ecDNA, first observed in cancer cells in 1962, has gained renewed interest due to its significant role in cancer. Recent studies show that 17.1% of cancer cells contain ecDNA, which is linked to worse clinical outcomes.
-
Role in cancer: ecDNA carries oncogenes and cis-regulatory elements that drive tumor growth and drug resistance. These DNA circles promote rapid transcription of oncogenes, contributing to aggressive cancer behavior and resistance to targeted therapies.
-
Formation and mechanisms: ecDNA forms through mechanisms like faulty DNA repair and chromothripsis. These circles can cluster together, enhancing transcription and promoting cancer cell survival and evolution.
-
Therapeutic and diagnostic potential: Researchers are exploring drugs targeting ecDNA to overcome cancer resistance. Additionally, ecDNA could serve as biomarkers for early cancer diagnosis, with efforts underway to develop diagnostic tools and therapeutic strategies.
This summary was generated by AI and checked by a human editor
It was back in 1962 that DNA circles were first spotted in cancer cells. ‘They called them double minutes, ’ says cancer biologist Paul Mischel from the University of Stanford, US. The ‘bunch of dots’ of DNA they saw in the microscope, separated from the chromosomes, but still within the cell nucleus, were often in pairs (now we know because they were replicating). Interest faded in what is now called extrachromosomal circle DNA (ecDNA), until the last decade. Armed with modern molecular biology and genomics tools, Mischel and others have started to realise their importance. These circles might explain why some cancers are more lethal than others.
Mischel had intended to focus his research career on precision medicine approaches to cancer – identifying the genetetic drivers of cancer by sequencing tumours and then finding targeted drugs. ‘But I was realising that there was a problem, something was missing. The precision strategy wasn’t working for a lot of people with cancer,’ says Mischel. He shifted his focus to studying these curious circles of DNA and began to understand their role in driving tumour evolution and drug resistance. In 2022, he received a $25 million (£20 million) Cancer Grand Challenges Award from CRUK and the National Cancer Institute, along with an interdisciplinary team from 13 institutions. Together, they hope to better understand these circles and ultimately find new classes of cancer therapeutics to target them.
Our cells are littered with circular DNA, alongside the expected chromosomes. ‘It seems like a natural part of life. You have seen them in all species we have looked at, in all tissues,’ says Anindya Dutta, a biochemist and cancer researcher at the University of Alabama, US, who studied these circles in mouse and human tissue. ‘I still remember my shock when the first experiment came in,’ says Dutta. He detected much smaller circles, the bulk of them are less than 2000 base pairs, which he calls micro DNA.
Mischel says these are not the same ones spotted back in the 1960s which are found exclusively in cancer and pre-cancerous cells and are considerably bigger, usually between one and five million base pairs and highly amplified. Earlier studies found these large circles in 1–2% of cancers, but the most recent study, based on data from almost 15,000 cancer patients with 39 tumour types, showed that 17.1% of cancer cells contained ecDNA. ‘We initially knew about glioblastoma and others, but we now know they are hugely prevalent in breast cancer and found in lung cancer, GI cancers, and many different cancer types,’ says Mischel. Comparing clinical outcomes showed patients who had ecDNA in their cancers did much worse than cancer patients without.
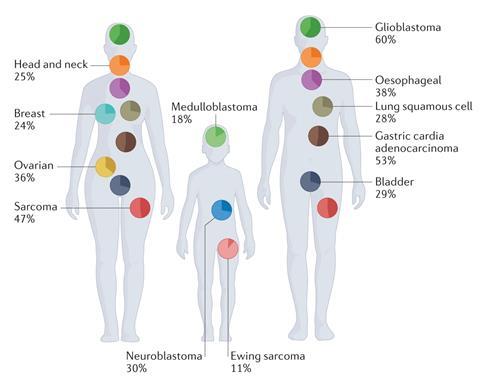
‘Now I think we’re beginning to understand why,’ says Mischel. These circles are large enough to contain genes and frequently carry oncogenes – the specific genes responsible for promoting cancer, which have shifted from the chromosomal DNA. ‘These ecDNA sometimes actually contain bits and pieces from different chromosomes within the same circle,’ says Anton Henssen, a peadiatric oncologist and researcher at the Max Delbrück Center for Molecular Medicine in Berlin, Germany, and part of Mischel’s eDyNAmiC grand challenge team. He had also spotted ecDNA in samples from relapsed neuroblastoma patients, a type of brain cancer only found in young children.
The circles also include non-coding regions of DNA known as cis-regulatory elements that bind transcription factors to promote the transcription of nearby genes. ’All of a sudden you have the possibility of interaction [between the genes and regulators],’ says Henssen. They have also found circles containing genes responsible for suppressing immune responses. Together these elements add up to the genetic recipe for driving tumor growth.
Henssen describes the structure of these circles as ‘dysregulated’. The DNA still forms chromatin – where the long strands of DNA molecules are coiled around histone protein complexes – but there are many loosely packed (euchromatin) areas, indicating active transcription. In fact, ‘they’re transcribing like crazy’, says Mischel; ‘you’re getting far more transcription of the oncogenes coming from these circles.’ Another fundamental difference from chromosomal DNA is that at the same time as copying themselves, ecDNA continues to be transcribed to produce RNA. This was ‘a shocking finding’, says Mischel. ‘Most genes are turned off during mitosis [cell division], but these oncogenes are not.’
Resistance and evolution
One of Mischel and colleagues’ earliest studies showed the link between ecDNA and resistance to cancer therapies. They found that drugs designed to switch off oncogenes reduced their presence on ecDNA, but they would quickly return once the drug was removed. Not unexpected, perhaps, ‘but the kinetics was all wrong, it happened much too quickly’, says Mischel. ‘You couldn’t explain that by classical genetics.’
They began to understand that when cells containing ecDNA divide, a different pattern of inheritance was taking place. In cell division, the linear chromosomes line up and copy themselves in an organised way before splitting so that each daughter cell has the same genetic material. ‘In ecDNA, these cell divisions are like a dice roll,’ explains Mischel. Random numbers of circles will be inherited by each daughter cell, some with lots, some with few. ‘What you get is a bell-shaped curve distribution, therefore you can select on the edges very quickly.’ So daughter cells with large numbers of cancer-promoting circles will preferentially survive and create a further generation of cells full of ecDNA-containing oncogenes.
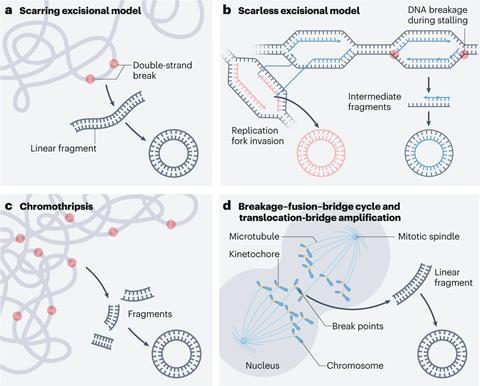
The increased variance and selection opportunity provides a super-charged mechanism for evolution. ‘That’s why these tumours are so resistant to the targeted therapies, because they actually change the number of the copies of the oncogenes so rapidly,’ says Mischel. The process can also explain how cancer cells can quickly switch the oncogene which drives a cancer, creating another way of resisting therapies – which Mischel says makes treating cancer ‘like a game of Whack-a-Mole’.
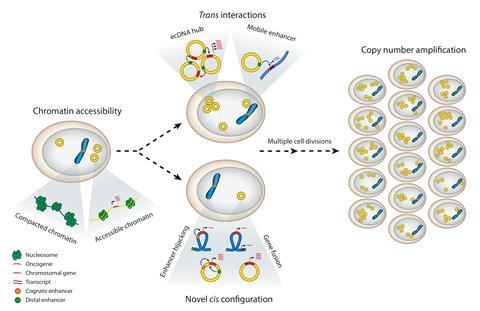
In 2021 members of the eDyNAmiC team discovered another piece of the puzzle – that different ecDNA circles in tumour cells cluster together in ‘hubs’ of 10 to 100 circles, allowing enhancer sequences to promote transcription between different circles. From studies of colorectal cancer cells, they found the circles were tethered together by an epigenetic reader protein which is able to recruit complexes that initiate transcription, increasing the over-expression of several oncogenes and making it likely these whole clusters would be segregated into the same daughter cell on division. Last year they further showed that if transcription was halted, this co-segregation of circles was reduced.
How these large DNA circles form in the first place is still being investigated. ‘They likely are generated by chance,’ says Henssen, but only those that are advantageous to the cell are perpetuated. Dutta says they seem to form when the mechanisms cells have for repairing DNA damage go wrong, particularly with double strand breaks. Dutta’s hypothesis is that when repairs fail, due to mutations in the repair genes, the cell cuts out the faulty DNA sections which then form circles. Another suggested mechanism is chromothripsis – which occurs when one chromosome gets separated from others in the cell cytoplasm, forming a micro nucleus. If its surrounding membrane becomes leaky and the DNA is exposed to nucleases, it shatters and the pieces reassemble into a circle. Mischel says their data suggests this contributes to 30–35% of ecDNA formation.
Smaller circles, detection and diagnostics
Similar mechanisms might be forming the circles that are present in all cells, but Henssen says ‘our view is that there are two distinct classes, the oncogenic ecDNA, and then all the other smaller circular DNA, and we believe that only those large ones that are amplified contribute to cancer.’ Dutta has looked at these other types of circles, of varying sizes and still sees suggestions of transcription. He used a sequencing method (ATAC-seq – assay for transposase-accessible chromatin with sequencing) that will only sequence DNA within open chromatin. The method only detects circles containing fragments originating from a single chromosome, but he has identified more than 18,000 of these mirco DNA circles in cancer cells, many carrying known cancer-driving genes.
‘Most circles cannot enhance the growth of tumors,’ says Birgitte Regenberg, a geneticist from the University of Copenhagen in Denmark, who has developed some of the foundational methods for studying extrachromosomal circule DNA. But her work has shown some smaller circles do play a role. ‘I know there’s a controversy about this, but I think that small circles can also provide selective advantages.’
We have shown small circles give tumor phenotypes like invasive growth and faster division
Her first studies of DNA circles were in yeast, where they are commonly used as an evolutionary mechanism. ‘If they provide a selective advantage, because their expression of the growth factor receptor EGFR leads to faster growth, you will have a fast accumulation of cells that carry a circle,’ she explains. In fact, unicellular yeasts could shift up to 20% of their genome onto these circles.
Regenberg thinks a similar mechanism is occurring in human cancer cells, be it the larger ecDNA or the smaller circles which she refers to as eccDNA (this young field is yet to standardise its terminology). Her lab has recently carried out studies (not yet published) showing the cancer-promoting role of smaller circles in colorectal and renal cancer cells. ‘We have reconstituted small circles in cell lines and shown that they give tumor phenotypes like invasive growth and faster division,’ she says.
One of Regenberg’s biggest contributions to the field so far has been pioneering a method for isolating circles of DNA from cells and disentangling the sequencing data obtained. This means removing the linear chromosomal DNA, as well as any mitochondrial DNA which is also circular. First, all DNA is immobilised onto magnetic beads and an enzyme will open up the mitochondrial DNA to make it linear. A nuclease then chops it up along with the rest of the linear chromosomal DNA, leaving just the circles.
The circles are then amplified and sequenced using a method called rolling circle amplification (RCA), which creates long single-stranded DNA molecules repeating the circle’s base sequences. Regenberg and colleagues developed the Circle Map algorithm to map these sequences onto identifiable parts of the genome by identifying the ‘joins’ in the circle that do not match known sequences. ‘We get a file out saying “This one came from this part [of a chromosome] or this circle came from this region of the genome,”’ she explains. Henssen says his group has adapted the protocol to sequence individual cells which has allowed them to detect the large differences in the number of circles in different cancer cells.
Regenberg is trying to use the method as more than an academic tool. She thinks the presence of circles of DNA could be a biomarker for early cancer diagnosis, or provide more information for treatment strategies. They have detected the circles in DNA purified from blood samples as well as tumour tissue. Dutta was also able to detect eccDNA in blood samples from mice and humans with lung and ovarian cancer. After tumours had been removed surgically, fewer circles were detected in the blood.
A significant proportion of the eccDNA Dutta detected was longer than 250 base pairs, which is longer than the circulating linear DNA found in blood. Linear circulating tumour DNA is already being analysed in cancer early detection tests or ‘liquid biopsies’. In 2020 Illumina spin-out company Grail partnered with NHS England to run a pilot study of their multi-cancer diagnostic test, involving 100,000 participants. Last year, NHS England reported interim results which they said had so far not been ‘compelling enough’ to justify further use of these tests.
Circular DNA is more stable in circulation, because it has no ends that can be easily degraded and Regenberg thinks it could provide an alternative diagnostic. She has been working with microfluidics expert Joerg Kutter at the University of Copenhagen to design a cheap lab-on-a-chip device that would provide a quick high-throughput automated method to purify extrachromosomal circle DNA ready for sequencing. The two enzymes that remove linear and mitochondrial DNA are immobilised on photo-polymerised emulsions inside microchannels, and the small size of the device speeds up all the reactions, making the process many times faster and able to remove over 99% of linear DNA .
The task now is to meaningfully interpret the results and find classifiers or biomarkers for cancer. There are still big challenges: circle DNA is present in very small amounts in the blood, plus there is the challenge of finding different profiles that identify different types of cancers. Regenberg has spun out a company and is starting to collaborate with Danish hospitals to analyse plasma samples from patients with pancreatic cancer, one of the deadliest forms on cancer with a five-year survival rate of only 13%. They are looking to find out if analysis of circle DNA can identify cancer recurrence earlier than CT scanning and optimise treatment.
Therapeutic strategies
Mischel and his collaborators are also looking for drugs that could target the larger ecDNA, and in 2018 co-founded Boundless Bio in California, US. Their first strategy focuses on the clash between transcription and replication occurring on the circles, which Mischel describes as like ‘two trains colliding’. ‘You have this massive amount of rapid transcription, [and] tons of nascent RNA being transcribed all the time, even when it’s trying to replicate,’ he says, which leads to a lot of damaged DNA. In most cells, regulation by kinases prevents these clashes. So Mischel’s team thought if they could stop the kinase working and cause even more transcription–replication conflicts, maybe cancer cells containing ecDNA will become overloaded with damaged DNA, leading to cell death. The strategy is ‘a little counterintuitive’ says Mischel: ‘We’re not trying to slow the cancer down – in a way we’re speeding it up.’ But disabling the cell brakes in this way will ultimately be lethal.
A genetic screen identified checkpoint kinase 1 (Chk1) as a target and Boundless Bio have discovered several molecules that act as inhibitors and kill cancer cells carrying ecDNA. Last year they also showed that, in mice with gastric cancer, they could prevent resistance to the drug infigratinib, another kinase inhibitor which shuts down the action of the growth factors that drive some cancers. Boundless Bio already has several drug candidate in clinical trials and is identifying other targets generated by ecDNA.
Another approach Mischel’s team are taking is looking at how ecDNA suppresses cell immune response; ‘We’re trying to understand how that happens and how to reverse it,’ he says. They are working in collaboration with chemist Ben Cravatt from Scripps Research in California, also a co-founder of Boundless Bio. He is bringing his pioneering activity-based protein profiling (ABPP) method to the problem of finding molecules that target the proteins involved in forming or sustaining ecDNA, which Mischel says are ‘the kinds of proteins that might not normally fall into traditionally druggable classes.’
‘ABPP allows you to, in a global way, identify small molecule ligands, for proteins in native biological systems, where they’re in their appropriate complexation states,’ says Cravatt. ‘It’s basically like a universal assay for all proteins in the cell.’ Cravatt uses a library of small molecules that have a reactive and usually electrophilic recognition group for targeting druggable sites. He then adds a broad-ranging chemical probe with a reporter group. ‘If any of the initial compounds you added is bound to a specific site, they block the broad probe from binding and the broad probe reactivity can be read out by mass spectrometry,’ Cravatt explains. ‘It’s a really nice way to look at tens of thousands of sites on proteins for interactions with small molecules in a single experiment.’
Henssen has also co-founded Econic Biosciences in London, UK, to find drugs that will stop ecDNA being maintained within cancer cells, although they have not made their strategy public yet. Dutta is also very hopeful that such drugs will help target currently undruggable oncogenes such as Myc, a regulator gene that codes for transcription factors, which he has found on circle DNA.
For Regenberg one of the key tasks to move the field forward is to try and create an atlas of the small and large circle DNA across the most common cancers to better understand the mutational signatures that correlates to each kind of circle and each type of cancer. ‘The atlas will show us the genetic features, [and] what the environmental features might be, like smoking or the impact of diabetes or obesity,’ she says.
‘I think we’re only at the beginning,’ says Mischel. Henssen agrees some exciting breakthroughs are on the way, ‘[The field] could make a big difference for cancer patients, if we do this correctly.’
Rachel Brazil is a science writer based in London, UK
Article updated 27 March 2025 to correct the spelling of Birgitte Regenberg’s name

















No comments yet