Protein-blocking chemicals better than genetic manipulation to uncover pathogen's infectious secrets
US scientists have used chemical inhibitors to block proteins that Chlamydia produces as it invades cells - because the bug’s infectious secrets can’t be uncovered by the traditional approach of manipulating its genes.
The ’chemical knockout’ technique, they say, may lead to new drug targets and help identify virulence factors in similar organisms where genetic strategies are difficult or nonexistent.
Chlamydia pneumoniae, a major cause of pneumonia, is hard to catch - at least in one sense. It can’t be grown on its own in the lab, reproducing only inside the cells of its hosts within storage compartments called vacuoles. The bacterium is therefore very well-protected from the kinds of genetic ’knockout’ experiments usually performed by researchers trying to analyse the functions of specific genes.
As making mutants isn’t an option in Chlamydia, Stephen Lory and colleagues at Harvard Medical School in Massachusetts focused instead on proteins released by the bacterium during its infection cycle. By producing them in yeast, they were able to establish that one in particular - CopN - was causing a block in the cell cycle, stopping yeast cells growing. When they transferred CopN to mammalian cells, they saw a similar effect.
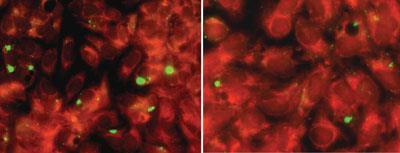
Unable to knock out the gene that produced CopN, the researchers used a high throughput screen of 40,000 compounds to identify small molecules that inhibited the bacterial protein and alleviated its disruptive effects on cells - effectively creating functional knockouts. The team thinks the molecules could also act as lead compounds for drug development.
Lory says he is surprised that the approach hasn’t been tried before. He thinks functional knockouts may have been perceived as less specific than genetic knockouts. ’I cannot ever guarantee that there are no secondary activities [in addition to blocking a particular protein],’ he says. ’But many genetic mutations have multiple effects in a cell too, so secondary consequences of mutations are not as certain as secondary activities of compounds.’
He says the same strategies could be used for other pathogens that are difficult to manipulate genetically, including the related bug responsible for sexually transmitted chlamydia infections (Chlamydia trachomatis) and Rickettsia species, which cause spotted fever and typhus. However, he stresses that these strategies could be just as important for complementing genetic studies in other organisms.
Ian Clarke, who studies Chlamydia at the University of Southampton, says the idea of using a library to screen for inhibitors is novel and could theoretically work for other microbes where no system is available for manipulating the genome. But he says a question mark remains over the Chlamydia strain the team used. ’It’s not the strain I would have chosen because it carries a bacteriophage. We don’t understand the relationship between the phage and chlamydia, so we don’t know what effect the phage might have on the chlamydial development cycle in the presence of the inhibitors.’
Hayley Birch
Enjoy this story? Spread the word using the ’tools’ menu on the left.
References
et alNature, 2008, DOI: 10.1038/nature07355




No comments yet