Novel proteins have been designed to neutralise some of the deadliest molecules in snake venom. The proteins, which were developed with the aid of deep learning, are more stable and easily produced than natural antibodies, and could potentially save many lives in the field.

More than two million people are bitten by snakes every year, resulting in over 100,000 deaths and 300,000 permanent disabilities, many in low-income countries. The cornerstone treatment is antivenom, which contains antibodies that bind the toxins. Today these are produced using animals. ‘You take the venom of interest, inject it into a horse and wait for a year or so for the immune system to generate antibodies to this small amount of venom,’ explains medical biotechnologist Timothy Jenkins of the Technical University of Denmark.
The resulting antivenoms are expensive and need refrigeration – which can preclude their easy availability in the field. Worse, they can contain few antibodies against some of the deadliest poisons such as three-finger toxins, which can be both neurotoxic and cytotoxic and are present in the venoms of around half the world’s deadliest snakes, including cobras and mambas. ‘The larger a protein the more likely the immune system is to create an immune reaction and since all of these three-finger toxins are quite small proteins the native immune response, in general, is bad at making effective neutralising antibodies,’ says Jenkins.
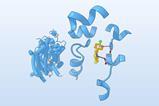
David Baker at the University of Washington in Seattle – who shared the 2024 chemistry Nobel prize – and his group teamed up with Jenkins and other researchers from the UK, US and Europe to design de novo proteins that would neutralise three-finger toxins. ‘Right now it’s a very standard pipeline that was developed in the Baker lab and is publicly available,’ says computational biotechnologist Susana Vázquez Torres, a PhD student in Baker’s lab. ‘The first step involves choosing a target… This algorithm explores different proteins that will bind to it – it’s like generating skeletons of proteins around your target.’ Next, the researchers used another program to select amino acid sequences that would maximise the protein–target binding. The final step used Deepmind’s AlphaFold to ascertain which ones would fold most suitably. After screening several hundred thousand candidates, the researchers synthesised the most promising ones.
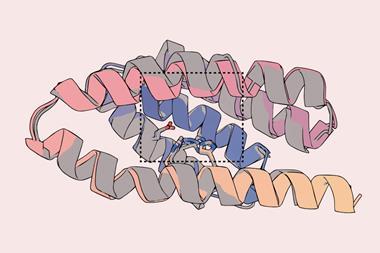
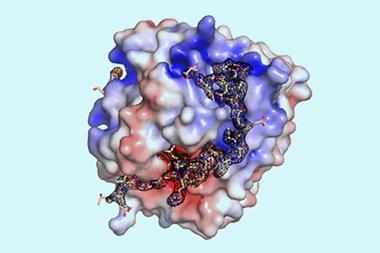
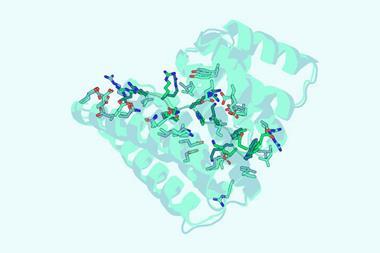
The small molecules they designed bind to the side chains of the three-finger toxins, sterically hindering their insertion into binding sites. For cytotoxic proteins, they hypothesised that this indirect approach might not prevent the proteins from disrupting lipid membrane formation, so they instead designed an unusually shaped protein that directly covered the tip. The resulting small molecules are thermally stable and can be produced using synthetic biology.
The researchers tested their antitoxin in several ways, including injecting mice with a cobra toxin before administering their antivenom up to 30 minutes later. ‘We had very low amounts of our therapeutic agent completely neutralise toxicity,’ says Jenkins. The researchers are now seeking to commercialise the technology, initially by augmenting existing antivenoms with small molecules targeting three-finger toxins, but they hope to later develop a standalone product.
Biological chemist Nicholas Polizzi at Harvard Medical School says that the work is ‘an obvious application of protein design but a very good one’. The half-life of small de novo proteins is normally hours rather than months, he notes, which could be an advantage or a disadvantage depending on whether the protein successfully binds the toxin before being excreted. Nevertheless, he says: ‘If you can use designed proteins to replace antibodies, you can imagine people carrying around a little “EpiPen” that’s designed for them to inject if they get bitten by a snake.’
Immunologist Joseph Jardine at the Scripps Research Institute in California, who has worked on monoclonal antibody-based antivenoms, agrees. ‘There’s not a lot of funding in this space, there’s not a lot of people working on it, and so all the tools that you can get that can try and solve it are going to be good.’
References
S Vázquez Torres et al, Nature, 2025, DOI: 10.1038/s41586-024-08393-x












No comments yet