Magnetic sorting method uses DNA to bind and release multiple targets
A new magnetic cell sorting technique uses principles borrowed from DNA nanodevices. The approach could help scientists rapidly separate different types of cells from complex mixtures.
Methods for separating diseased cells from healthy ones form the basis of certain diagnostic approaches. Sorting techniques are also needed to study immune responses and other complex biological systems. Magnetic sorting is well established, but when several types of cell or other target are involved, the process becomes complicated and longwinded.
Christine Probst at the University of Washington in Seattle, US, explains that her team’s method, which is designed to sort lots of different targets simultaneously, is based on a principle called ’strand displacement’, better known to those in the field of DNA nanotechnology. ’It was one of those crazy ideas that we weren’t sure was going to work,’ she says. ’We saw a connection and we thought this could be really high impact if it works. And it did work astonishingly well.’
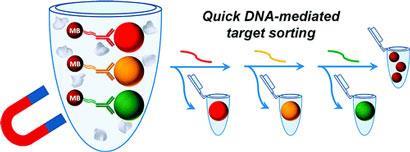
Strand displacement exploits the fact that a pair of DNA strands - complementary strands - can be unzipped by the introduction of another longer, complementary strand. Molecular engineers use this zipping and unzipping to drive molecular switches and motors. But the team realised the displacement effect could also be applied to release cells sequentially, to sort them into groups.
Their study provides proof of principle using coloured beads. Each colour, bearing a different immune protein on its surface, is released in a separate round of displacement and then captured. The initial strand pairing is formed between DNA linked to an antibody, which recognises the immune protein on the coloured bead, and a complementary DNA strand attached to a second, magnetic bead. When a third strand arrives to break up the pairing, the coloured bead is released and the non-magnetic portion of the sample is collected.
The whole process takes less than a couple of hours, as opposed to five for comparable approaches, and produces samples that are more than 95 per cent pure. Although the purity of collected beads was reduced when one target was rare, Probst says the process can be optimised by designing DNA sequences that bind more securely.
The technique is clever, and faster than most documented in the literature thus far, says Jeffrey Chalmers, who works on cell separation at Ohio State University in Columbus, US. But that doesn’t necessarily mean it will be easily applied. ’Each system you work with has its own, unique challenges,’ he says. ’Time will tell, as this approach is applied to other systems, how it will work.’
Hayley Birch
References
C Probst et al, J. Am. Chem. Soc., 2011, DOI: 10.1021/ja2072324




No comments yet