First bacterium with a totally redesigned genome created
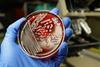
Designer E. coli uses a simplified genetic code to make proteins
A bacterium has been created with a totally synthetic genome that can also produce proteins using fewer triplet DNA bases – or codons – to encode the standard amino acids required for life.1 The discovery could serve as a platform for developing new proteins with unique properties.