A new generative AI model rapidly generates diverse antimicrobial peptide structures for screening against treatment-resistant microbes. Subsequent in vivo studies identified two promising lead candidates for further clinical development and the team behind the work believe this approach could accelerate drug discovery in both peptides and small molecules.
Antibiotic resistance is one of the biggest threats to the future of human health, with drug-resistant infections predicted to surpass cancer as the leading cause of death by 2050. In recent years, antimicrobial peptides (AMPs) – short chains of between 10 and 50 amino acids – have emerged as a promising solution to this resistance problem. ‘AMPs target bacterial membranes,’ explains Jon Stokes, an antimicrobial chemical biologist at McMaster University in Canada. ‘If you have a molecule that is biophysically disrupting a cell envelope, resistance is much harder to evolve.’
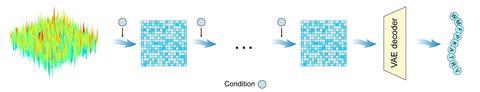
However, navigating the vast chemical space of potential AMP structures is an overwhelming challenge. AI tools have proven a valuable method to streamline this process, but the structures generated by existing systems often lack diversity as the models struggle to extrapolate beyond the training data.
Now, using a more complex type of generative AI system, Wenqiang Chang and his team at the Shandong University in China have developed an effective tool to rapidly design new and diverse AMPs with varied antimicrobial activity. ‘Our pipeline leverages a latent diffusion model, which operates within compressed data representations,’ explains Chang.
The model works by compressing the training data of 2.8 million peptides into numerical form – known as latent space – a process achieved by amplifying the signal noise of each data entry randomly. The system then extrapolates new peptides out of these simplified data, stripping away the noise to decompress each result back into a peptide sequence. ‘The denoising process is stochastic, meaning the model does not always remove noise in the exact same way,’ explains Stokes, who was not involved in the project. ‘This small randomness in denoising trajectories ensures diversity in generated peptides.’
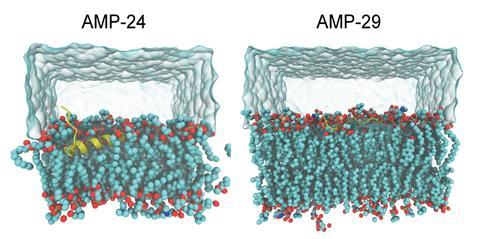
The generated sequences were then immediately filtered through a classification pipeline to identify the hits most likely to possess antimicrobial properties. Of the 600,000 results, Chang’s team experimentally tested 40 against fungal and bacterial pathogens, with 25 exhibiting antimicrobial properties. Safety profiles of the most potent peptides identified two candidates with potential for in vivo use and antibacterial AMP-24 and antifungal AMP-29 were successfully used to treat microbial infections in mice.
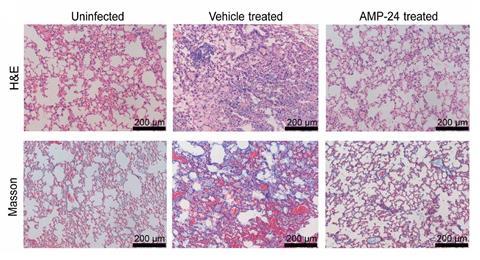
Stokes was impressed by the team’s thorough approach and is excited to see how this technology pipeline could influence drug discovery in areas beyond antimicrobial peptides. ‘The fact that they were able to generate peptides that show in vivo potential and have reasonable toxicity profiles is super promising and I’m sure there’s going to be a lot of valuable learning that can be gleaned from this,’ he says. ‘An important next step is determining whether AMP-24 or a derivative thereof is progressible as therapeutic. And in terms of the model, one development that I really want to see is the incorporation of toxicity filters really early to keep this end goal in focus.’
Chang’s team are already exploring advancing AMP-24 towards the clinic and plan to refine the model further to improve the peptides’ properties across multiple parameters, including cytotoxicity. While their next big focus will be on expanding the pipeline to identify multifunctional antimicrobial peptides, the team hope that researchers in other areas of drug discovery will leverage the method towards different targets. ‘[The model’s] core value lies in accelerating drug discovery [and] lowering development costs, potentially transforming traditional molecular discovery paradigms,’ says Chang.
References
Y Wang et al, Sci. Adv., 2025, DOI: 10.1126/sciadv.adp7171













No comments yet