Oxford Nanopore sequences a viral genome and aims to launch its sequencing platforms within the year
Oxford Nanopore has presented the first complete genome to be sequenced using a nanopore. The UK company says this small viral genome is its first step towards sequencing a complete human genome in 15 minutes and developing disposable USB drive sequencers.
The inexorable race for faster, cheaper and more accurate DNA sequencing, driven by the promise of personalised healthcare, has spawned numerous different technologies for improving the process. Oxford Nanopore’s ‘strand sequencing’ devices are the latest to aim for a slice of the highly lucrative market.
It is the company’s nanopore technology that differentiates it from competitors. Rather than using fluorescent labels on the four different DNA bases, the sequence is read by monitoring an electrical signal as a strand of DNA passes through a mutant variant of the pore protein, alpha-hemolysin. Each base gives a subtly different reading as it passes through the pore, allowing direct reading of the sequence without generating vast amounts of complex optical fluorescence data.
One of the biggest advantages of the technology is the read length, explains Oxford Nanopore’s Zoe McDougall. Current machines can typically only handle fragments of DNA a few hundred base pairs long, she says. ‘We’re showing a genome of around 5000 base pairs and we’ve sequenced it as a hairpin - we’ve sequenced both strands in one go.’ That gives a read length of around 10,000 bases, but McDougall adds that they have demonstrated read lengths of 48,000 bases and are aiming to get up to 100,000.
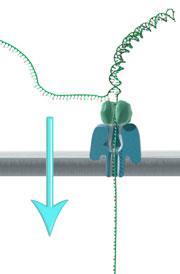
Part of the challenge in developing this technology has been controlling how the DNA strand moves through the nanopore, but this has been addressed using a ‘processive’ enzyme attached to the pore - again engineered from a natural version by the company to suit its needs. It is the processive enzyme that binds the DNA, allowing it to dock into the nanopore, and then ratchet through.
McDougall points out that the there is a balance between speed and accuracy. The faster the DNA goes through the pore, the noisier the signal. ‘But the area where we’re detecting the signal is a few bases long,’ she adds. Each time the strand moves along you get a different combination that can be distinguished by the company’s algorithms, so each base is effectively double-checked a number of times to improve accuracy. She notes that the error rate in reading is still higher than the company would like, at around 4%, but work is ongoing to bring that down.
John Milton, Oxford Nanopore’s chief scientific officer explains that the company is aiming to have two versions of the machine on the market this year. One is a modular system called GridION, which can be set up as anything from a single benchtop sequencer to a workhorse with multiple units in parallel, which the company projects could eventually sequence whole human genomes in a matter of minutes. The second is the MinION, a miniature, self-contained disposable sequencer that interfaces with a computer via USB.
‘The sequencing market is currently very competitive and rapidly changing, and the release of an instrument from Oxford Nanopore has been widely anticipated,’ says Julian Parkhill, senior group leader at the Sanger Institute in Cambridge, UK. He explains that uptake of the new machine by small and large users will depend very much on the details of the operation, such as price per base pair, speed and accuracy. ‘We will want to carefully evaluate any new technology and see what advantages it may have over existing systems in these areas.’
Milton agrees that this kind of evaluation is a key part of the launch process. ‘We’ll be working closely with the big sequencing centres,’ he says. They’re interested in the workflow and details of what the data look like, which helps fine tune the process. ‘To start with we invite them to come and run their samples on our machines. Then we take machines out to them with our employees running them, then transition to running it themselves. It’s a very controlled process.’
















No comments yet