Synthetic ‘enzyme’ matches natural counterparts for catalytic power
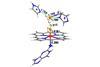
Nanoparticle, nucleic acid and peptide trio mimics peroxidase enzymes
A nanoparticle with a catalytic turnover on a par with a biological enzyme has been constructed by researchers in China. Such an entity takes us one step closer to materials with the catalytic power of proteins without their drawbacks or complexity.
