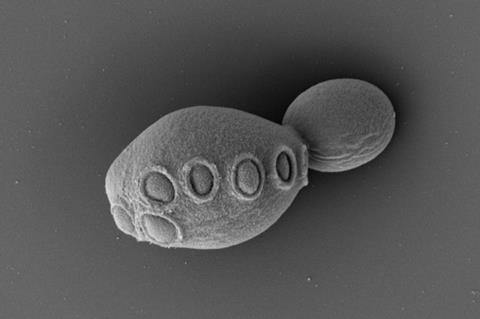
A strain of yeast that has a genome that is more than 50% synthetic DNA has been produced in the lab for the first time. This is the latest milestone in a 15-year effort to design and build an entirely synthetic eukaryotic genome from scratch that the team hopes to finish within the year.
The synthetic yeast genome project, also known as Saccharomyces cerevisiae (Sc) 2.0, comprises research groups from around the world who have each taken ownership over producing a synthetic version of one of the 16 chromosomes that make up the yeast genome.
‘We have about 10 teams from top universities from four continents; it’s a truly international effort,’ explains Patrick Cai, chair in synthetic genomics at the University of Manchester and international coordinator of the Sc2.0 project.
Ben Blount at the University of Nottingham and the leader of one of the UK-based teams explains that yeast has been used as a model organism for eukaryotes for many years, meaning scientists likely know more about yeast than any other organism. ‘The yeast genome is really nice and compact,’ he explains. ‘It also has this impressive innate ability to just stitch DNA together. So, if you have two DNA sequences that have a similar sequence between them, the yeast will just stitch that DNA together.’
A package of eight papers was recently published across Cell, Molecular Cell and Cell Genomics in which teams within the Sc2.0 consortium described their synthetic yeast chromosomes and highlighted key features they had identified. In addition, the consortium published a proof-of-concept study in which 6.5 of the synthetic chromosomes, plus an extra ‘neochromosome’ consisting only of transfer RNA (tRNA) genes, were brought together into a single cell that was able to survive and replicate in a similar way to wild yeast.
‘We show we have the technology to merge over 50% of the synthetic DNA in one single cell,’ explains Cai. ‘This is a landmark in biology and biotechnology – this is the most synthetic organism we have ever made.’
The design of every chromosome was carefully controlled by a set of strict design principles, explains Blount, whose team was responsible for chromosome XI, before an extensive and lengthy ‘debugging’ process. However, rather than being a straight copy of the natural genome, the Sc2.0 synthetic genome contains thousands of designer edits, including deletion of mobile elements and introns, relocation of transfer RNAs (tRNAs), and swapping of stop codons to increase genome stability and generate a variety of phenotypes with desirable properties, such as immunity to viruses.
Blount’s team also showed that its chromosome could be repurposed as a new system to study extrachromosomal circular DNAs. These free-floating circles of DNA that loop out of the genome are increasingly recognised for their role in ageing and in cancer.2 ‘These circles are ways that tumours can make themselves resistant to a lot of the treatments that we use,’ says Blount.
Cai’s lab also created an entirely synthetic ‘neochromosome’, made solely from 275 tRNA genes relocated from the original chromosomes.3 ‘Nothing like this exists in nature,’ says Cai. ‘[But] we installed it in the yeast [cell], and it seems to be happy with it, which means it’s functional.’
Now the process is underway to bring each of the chromosomes together within a single yeast cell and it is expected that the complete synthetic genome should be finished within the year.
John Glass, leader of the synthetic biology group at the J. Craig Venter Institute where a team produced a bacterial cell with a synthetic genome in 2010, says he sees the Sc2.0 work as ‘a continuum’. ‘Every new step as they advance closer to their ambition of making a yeast cell that has a completely synthetic genome, they are finding extraordinary and unexpected things about eukaryotic biology that are fascinating and, over time, will have extraordinary consequences for medicine, for industry, for our view of ourselves,’ he explains.
‘They have kept this group of at least 16 different academic and industrial and government organisations together for more than a decade and kept it funded through a huge variety of sources. And this is not a trivial undertaking,’ he adds.
References
1 Y Zhao et al, Cell, 2023, 186, 1 (DOI: 10.1016/j.cell.2023.09.025)
2 BA Blount et al, Cell Genom., 2023, 3, 100418 (DOI: 10.1016/j.xgen.2023.100418)
3 D Schindler et al, Cell, 2023, 186, 1 (DOI: 10.1016/j.cell.2023.10.015)















No comments yet