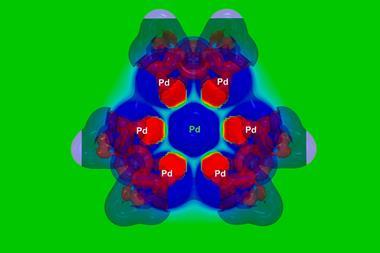
Combining machine learning with computationally derived descriptors has allowed scientists to find new examples of a special class of catalyst using only a few experimental data points. The team led by Franziska Schoenebeck from RWTH Aachen University in Germany developed a workflow that identified 21 phosphine ligands that may form dinuclear palladium(I) complexes with a certain geometry and air stability over the more common palladium(0) and palladium(II) species.1
‘These dimers are very promising catalysts with distinct reactivity relative to most of the commonly used palladium-based catalysts,’ comments Tobias Gensch from TU Berlin, Germany, who was not involved in the study. ‘However, their chemistry is not yet well understood because their synthesis was unpredictable and the influence of the ligand on the dimer stability was unknown.’ The new approach allowed the researchers to predict ligands that stabilise palladium(I) dimers and synthesise several new examples of these complexes, he says.
Discovering efficient catalysts is key to many innovations in chemistry, but different species have different activities and selectivities, so finding the right compounds is challenging. ‘Accurately predicting the speciation of a catalyst would, in principle, require precise knowledge of all the species that can be formed under given conditions and their relative energies – a daunting task!’ points out Marc-Etienne Moret, an organometallic chemist at Utrecht University in the Netherlands.
That’s why chemists usually rely on trial and error, testing ligands that they think could work. ‘Scientists have also developed maps to classify the ligands according to their properties; this can help them identify promising candidates visually,’ Moret adds. But in some cases, these methods don’t help. The new results show that machine learning can successfully predict ligands where neither intuition nor visual inspection would succeed, he says. ‘This could accelerate the development of new catalysts by identifying promising targets before making and extensively testing them in the lab.’
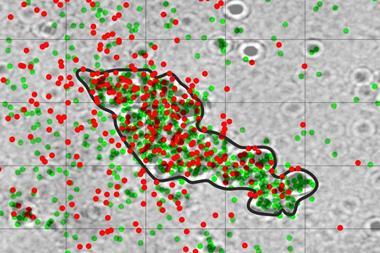
The scientists first used an algorithm to filter 348 ligands based on their general properties and then carried out additional clustering by introducing problem-specific data obtained from density functional theory calculations. This strategy allowed them to group a large data set into smaller subsets of greater similarity, tailored to the problem at hand. The team then verified some of the predicted ligands experimentally, including one that had never been synthesised before, and used them to make new palladium(I) dimers.
Gensch notes that the system could identify novel ligands using only five experimental data points. ‘Other machine-learning approaches such as regression modelling require far more data as input,’ he says. ‘The ability to work with so little data results from the combined use of a general-purpose ligand database and highly informative problem-specific descriptors, paired with a simple, yet powerful, two-stage clustering approach.’
‘The accuracy of the algorithm’s predictions is remarkable,’ Moret says. ‘It suggested ligands that would otherwise probably never have been tested. This methodology could potentially help to solve many related problems for which empirical or computational data exists but does not yet form an intuitively understandable picture.’
References
1 JA Hueffel et al, Science, 2021, DOI: 10.1126/science.abj0999










No comments yet