Fungal infections account for more than 1.5 million deaths globally each year; more than malaria and breast cancer. However, therapeutic options are limited and resistance to the small number of antifungals available is growing. Despite this, fungal infections remain in the shadow of bacterial infections as a major contributor to antimicrobial resistance (AMR) and progress to develop new therapeutics has been slow, in large part due to the limited funding available.

One lab that has placed fungal pathogens front and centre of its research is the Cowen Lab in Toronto, Canada. Set up by Leah Cowen – a mycologist – in 2007, the lab is driven by a desire to answer the big questions surrounding fungal drug resistance, development and disease, namely, how do these microbes cause disease and how do they evolve resistance? ‘We have very collaboratively built out a strategy to leverage genomics to understand vulnerabilities in fungal pathogens, and also chemical biology to identify molecules that we might be able to harness to thwart them,’ explains Cowen.
As Cowen has gradually taken on additional responsibilities outside the lab, she has built a strong leadership team to help her within it, including senior research associate Nicole Robbins, a previous graduate of Cowen’s who plays a significant role in mentoring students. ‘I always envisioned working in an academic lab and working with students and helping them with their careers and their training,’ says Robbins.
Another senior research associate is Luke Whitesell, a paediatric oncologist by training, who brings substantial expertise in pharmacology. ‘I have been in multiple labs over the years [but] this is the most nurturing environment I’ve ever seen and by that, I don’t mean soft – there’s some tough love that goes on – but there’s such an emphasis on the development of each individual.’
The lab’s programme of work runs across six themes: functional genomics, chemical genomics, mechanisms of drug resistance and disease, the fungal microbiome, structure-guided drug design, and the evolution of resistance. ‘We have lots of different scales of projects [including] some very large-scale projects where we’re building out a mutant collection, for example, to be able to control expression of every gene in a particular pathogen’s genome; that’s a huge labour of love,’ says Cowen.
A new antifungal compound
Treatment of systemic fungal infections is currently limited to three major classes of antifungals: azoles, such as fluconazole; echinocandins, such as caspofungin; and polyenes, such as amphotericin B; each of which target components of the fungal cell membrane or cell wall. Unfortunately, each of these antifungal classes has significant drawbacks and their overuse in clinical settings has led to a surge in resistant isolates.1
One particular strategy being trialled is combination therapy; combining compounds has the potential to increase efficacy as well as slow the evolution of drug resistance.

To explore this approach, the Cowen Lab teamed up with a group of scientists at the Riken Center for Sustainable Resource Science, in Japan. Together they screened over 20,000 natural products and synthetic small molecules for antifungal activity against Candida albicans – among the most prevalent of the fungal species – and three other fungal pathogens; Aspergillus fumigatus, Cryptococcus neoformans and the emerging pathogen, Candida auris.
‘Two of our fantastic graduate students travelled all the way to Japan to do this initial screen,’ explains Robbins. ‘They looked not only for compounds that had activity on their own, but compounds that enhanced the activity of fluconazole in these primary screens. Then, once they had returned, there was a whole series of triaging steps; looking for potency and fairly minimal, mammalian cytotoxicity.’
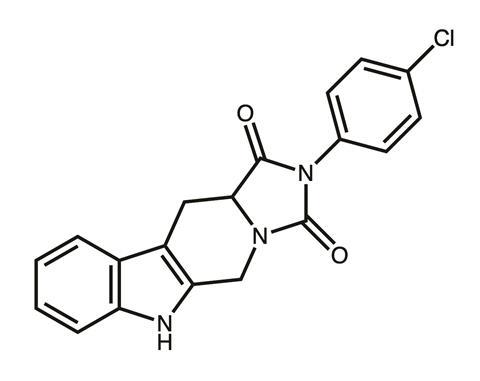
Eventually the team honed in on a particularly potent imidazopyrazoindole compound, NPD827, which substantially increased the activity of fluconazole against azole-sensitive strains and azole-resistant strains of C. albicans.2 However, pinning down the previously undescribed mechanism of action was a significant challenge and required just about every tool in the lab’s armoury.
Mechanism of action
‘It was a journey, it took a really long time,’ says Robbins. ‘We tried to use a lot of our chemical-genetic methods – selection for resistant mutants, looking for hypersensitivity in the genomic deletion collections – we would get little hints here and there that it probably had something to do with the membrane, but it was becoming more and more apparent, the harder we tried, that this was not a single protein target.’
After much brainstorming and collaboration, the team found that NPD827 was directly binding to the sterol-containing lipid bilayers of the fungal membrane and, within minutes, inducing a profound effect on membrane homeostasis and fluidity, particularly in the presence of sterol-biosynthesis-perturbing agents, such as fluconazole.
This disruption triggered a whole host of downstream ramifications by activating key membrane-associated stress responses to cope with the imbalance, such as accumulation of lipid-storage organelles, known as lipid droplets, induction of the unfolded-protein response, and activation of calcineurin-dependent stress responses. ‘That’s what’s causing [the pathogen] to become hypersensitive to the azoles,’ explains Robbins. ‘It was a really tricky, but really unique mode of action that we ended up uncovering through a variety of genetic, biophysical and biochemical assays,’ she adds.
They also discovered that NPD827 caused the drug-efflux pumps situated in the C. albicans plasma membrane to become non-functional. ‘[NPD827] doesn’t clog up the pump – it does not act as a competitive inhibitor – it actually causes one of the major efflux pumps in C. albicans – Cdr1 – to accumulate,’ explains Whitesell. ‘So, you see more of the pump at the plasma membrane, but it doesn’t work. The consequence is that at the same time as causing a lot of stress on the organism, you’re making the azole more effective because it accumulates better within the cell.’
Following this exciting discovery, the team partnered with additional collaborators to explore the therapeutic potential of using NPD827 as a way to reverse azole resistance in C. albicans and inhibit key virulence traits, including filamentation and biofilm formation.
‘C. albicans can grow in a circular yeast-like state, as well as growing in polarised projections or filaments,’ explains Robbins. ‘But in vitro we noticed that NPD827 was able to block this transition in response to a few filament-inducing cues.’ One of the things that filamentation is really important for is biofilm formation: ‘the yeast tend to adhere to a surface – in patients, this might be a catheter – and then the biofilm develops by C. albicans forming both yeast and filaments, in order to form a complex community.’
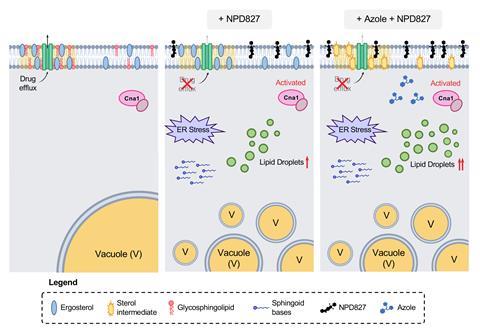
Although NPD827 does not have the necessary pharmacological properties for systemic use (they are still in the process of defining the specific issues), the team was able to demonstrate that when combining it with fluconazole it did act in a fungal-selective manner in a co-culture model with human cells. And likewise, in a Caenorhabditis elegans model it improved survival when the worms were infected with azole-tolerant and -resistant clinical isolates.
The team also reached out to a collaborator who was able to test what they had seen in a rat catheter model of C. albicans biofilm infection to find out if NPD827 was also able to block biofilm formation in a central venous catheter similar to the ones used in human patients, and it did. ‘We saw a significant reduction in colony forming units of C. albicans that was grown within the catheter,’ Robbins adds. ‘Not only that, but when we looked at how robust the biofilm was, we noticed that it wasn’t very strong; as soon as we tried to image the catheter, it looked completely sterile.’ This suggests that NPD827 has great therapeutic potential in terms of abrogating biofilm formation in this particular mammalian model.
Potential for new therapeutics
The work that the Cowen Lab has done in this area highlights that there are a whole host of chemical compounds that could have great efficacy against fungal pathogens but are yet to be explored. ‘The fact that we found so many compounds that not only worked alone but worked in combination with the azoles right then and there, [it shows] there’s lots of promise for developing new therapeutics,’ says Robbins.
‘The idea of being able to take the most widely-deployed class of antifungals, the azoles, and restore their efficacy, is a really neat idea,’ Whitesell adds. ‘We’ve spent a lot of time and effort developing azoles over the last few decades, and if we can restore their efficacy that could have a very broad and immediate impact on clinical application.’
It’s incredibly collaborative
Of course, there are still problems to overcome; for example, coming up with a way to translate the preclinical findings with NPD827 into a useable therapeutic, but Whitesell is optimistic: ‘There’s lots of chemical tricks to attempt; a prodrug approach could be highly effective,’ he says.
At the Cowen lab, a significant emphasis is placed on mentoring and on building the next generation of scientists to ensure that they can extend and expand this important work. Currently, the lab has over 20 trainees, spanning postdocs, research associates, graduate students and undergraduates, but despite its size, the lab has remained a nurturing environment.
‘Leah’s vision of making sure everybody gets personalised mentorship, and making sure she has a really strong leadership team in place gives it a bit of a small lab feel,’ says Robbins. ‘Not only do students get lots of support from senior members of the team, but it’s also incredibly collaborative.’ Whitesell adds: ‘what sets the lab apart is this emphasis on mentoring, and rather than an exploitation of people to get stuff done. It’s really a developmental process; it’s been a joy for me to be associated with the effort.’
Extensive collaboration also happens outside the lab, as demonstrated in the work on NPD827, to ensure that the very best minds are brought together to provide answers to the most critical of questions around antimicrobial resistance. But to continue making progress, more resources and funding are urgently needed, says Cowen. ‘We’re really strong in our field with the early-stage discovery pipeline; we have lots of really cool targets and lots of interesting molecules, but there’s a need for additional investment and resources to be able to drive the next stages as you advance molecules through a pipeline towards the development strategy.’
References
1 KR Iyer, N Robbins and LE Cowen, iScience, 2022, 25, 103953 (DOI: 10.1016/j.isci.2022.103953)
2 NM Revie et al, Nat. Commun., 2022, 13, 3634 (DOI: 10.1038/s41467-022-31308-1)















No comments yet