IBM model can also find molecules that bind at different sites on target proteins
An artificial intelligence (AI) model has identified potential antiviral molecules that inhibit the Sars-CoV-2 virus, from only the sequence information of target proteins – without any knowledge of the proteins’ 3D structure or known binding sites.
Researchers from IBM and the University of Oxford, UK, trained a generative AI called CogMol, part of IBM’s MolFormer-XL model, using general information about proteins and their binding properties. They then used this model to identify potential antivirals for the Covid-19 virus. The pool of suggestions was narrowed down using IBM’s RXN neural network to identify those that could potentially be synthesized. The most promising were then made by contract manufacturer Enamine in Ukraine, and tested against the virus. ‘[It took] only a few weeks with the minimal amount of knowledge to come up with new inhibitors for a novel pathogen,’ explains Payel Das, manager of IBM’s AI science department.
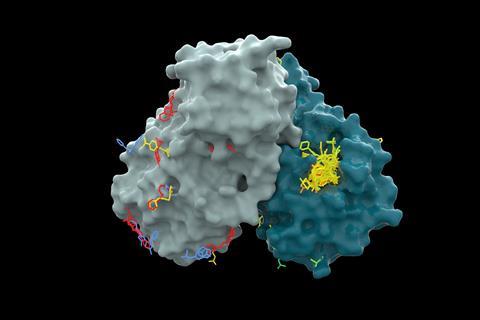
Combining CogMol with other AI algorithms allowed the team to focus the thousands of initial suggestions towards molecules with more drug-like properties, as well as their ability to be quickly synthesised. Chemists at Enamine then selected eight to make – four directed at each of the spike and main protease. In laboratory tests, two from each set of four successfully inhibited their respective protein targets. Das suggests that even this modest 50% success rate – given the speed and simplicity of the models inputs – means that ‘this sort of technology can help humans to explore the chemical world’ and rapidly assess various possibilities. ‘What you also find is that this kind of study can identify new mechanisms of targeting this novel pathogen.’
The IBM researchers prompted CogMol with the protein sequences of the Sars-CoV-2 spike and main protease proteins to try to pair with possible antivirals. While other AI programs need structural data or representations of molecules to predict their properties, CogMol’s inputs are simple text strings relating only to the protein’s sequence. ‘The method makes no structural predictions, neither does it draw inferences about binding pockets or other physical motifs,’ says Jason Crain, an IBM senior research scientist and visiting professor at Oxford.
As well as the molecules that were selected for synthesis, the team analysed many of CogMol’s other suggestions computationally, to determine where and how they might dock into structural models of the target proteins. They found molecules that could bind to relatively unstudied sites on the spike protein. ‘We were surprised to know that the molecules do not attack [the usual] target spots, the [receptor binding domain] of the spike protein,’ Das adds. ‘We didn’t even know that would have been possible.’
As Das and Crain explain, alternative binding sites for antivirals – away from where the protein is binding its biological target – might be less susceptible to mutations as the virus evolves, which could reduce the need for new antivirals to keep up with the mutations. The researchers hope to proceed with laboratory trials for some of these molecules to explore their antiviral abilities.
As Oxford established its Pandemic Sciences Institute in 2021, Das, Crain, and the other IBM researchers are hopeful to apply CogMol and its umbrella software MoLFormer-XL to other viruses as part of this more extensive initiative. ‘[The Institute] is geared up to create this strongly disciplined bridging environment where you have AI technology, and physical modeling, all focused together,’ Crain says. ‘This [study] was an agenda setter [for future collaborations].’
IBM has already established a partnership with pharmaceutical firm Moderna in April 2023. Moderna is looking for new antivirals for Severe Acute Respir Sars and other viruses, so using IBM’s CogMol software could accelerate this process. ‘With Moderna, we are using this entire suite of MolFormer, including CogMol to help them design new RNA vaccine stabilisers,’ adds Das.
References
V Chenthamarakshan et al, Sci. Adv., 2023, DOI: 10.1126/sciadv.adg7865












No comments yet