An organic synthesis algorithm has mapped out the thousands of reactions that might have converted abiotic compounds into the building blocks of life more than 3.5 billion years ago. Starting off with six simple precursors, the program discovered many known as well as 24 entirely new pathways to prebiotic molecules, and showed how catalytic and self-replicating systems might emerge.
Despite hundreds of demonstrations that various organic reactions can take place under the conditions on early Earth, the scientific community still only has a piecemeal understanding of how the building blocks of life emerged. That’s because the number of possible combinations of these reactions is so large that the number of molecules generated quickly jumps into the tens of thousands. While synthesising and analysing so many compounds is difficult, it could in principle be sorted using a computer.
Now, researchers have done just that. A team led by Bartosz Grzybowski and Sara Szymkuć from the Polish Academy of Sciences encoded all 500 known prebiotic reactions and a feedstock of six precursors – water, hydrogen cyanide, ammonia, hydrogen sulfide, nitrogen and methane – into open-use platform Allchemy. The algorithm then used encoded mechanistic chemistry rules to produce a map of their combinations.
Running the program for seven generations, each time combining the generated molecules with what came before, the researchers ended up with almost 35,000 compounds including 50 biotic ones. The program was able to find many prebiotic syntheses previously described in the literature, for example 10 pathways leading to the DNA component adenine. But it also discovered 24 entirely new pathways to biotic compounds – more than 20 of which the team experimentally validated.
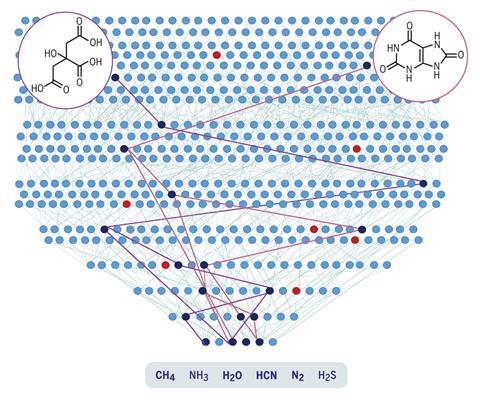
As well as that, more complex systems such as reaction cycles and micelles began to emerge. One molecule generated, iminodiacetic acid, can complex with metals such as manganese to act as a catalyst. ‘You can take it through a cycle and it can potentially reproduce two copies of itself for one cycle. In experiment, we showed this auto-amplification. What this teaches us is that this self-replication emerges chemically,’ says Grzybowski.
One of the reasons computer programs are better at finding these pathways than humans, explains Szymkuć, is that ‘people aren’t accustomed, when designing something, to incorporate a step, which degrades a molecule, and that’s necessary for the cycle.’
But the vast majority of reaction combinations led to abiotic compounds – molecules that were never incorporated into living systems. Analysis showed that the molecules that became the building blocks of life were more soluble in water and more thermodynamically stable. ‘The chosen ones had a balanced number of hydrogen bond donors and acceptors,’ Grzybowski adds. ‘When you think about it, this makes perfect sense – think about DNA. Nature chose molecules that are more likely to be fitting into a larger architecture, to build these kinds of supramolecular assemblies.’
‘This is an incredible piece of work,’ says Valentina Erastova who investigates origin of life chemistry with computational methods at the University of Edinburgh, UK. ‘We have a bit of a limitation in that this has to be based upon our discovered reactions. There are still reactions we haven’t been exploring as much that will be related to minerals and surfaces, which could be catalytic. That would be very interesting to look into… we can’t take this map as an absolute map.’
References
A Wolos et al, Science, 2020, 369, eaaw1955 (DOI: 10.1126/science.aaw1955)










2 readers' comments