
A hand-powered centrifuge has been combined with an inexpensive assay to detect Sars-CoV-2 in saliva samples. The setup was invented in the lab of Manu Prakash at Stanford University and aims to make diagnosis of the disease possible in remote or resource-limited regions.1
‘Most conventional tests [for Covid-19] so far are focused on high throughput, large-scale detection in metropolitan hospitals,’ says Prakash. His diagnostic setup can be built with readily available components, costing less than $5 (£3.90) per unit. The reagents cost around a dollar per test.
He has begun to validate the device in the US and Bolivia, partnering with a community of doctors in the Amazon region serving people in remote locations. ‘Signatures of the virus are spreading in indigenous communities, which have no healthcare infrastructure to handle something like this,’ says Prakash.
The process relies on an assay developed by Constance Cepko and her student Brian Rabe at Harvard University and Howard Hughes Medical Institute that is based on loop-mediated isothermal amplification (Lamp). This is capable of amplifying microgram quantities of viral RNA at a single reaction temperature.2
(1/n) Excited to share Handyfuge-LAMP; a low-cost & electricity-free centrifugation for isothermal SARS-CoV-2 detection in saliva. With COVID19 cases increasing globally, diagnostics needs to be accessible to everyone - including resource limited regions https://t.co/3T5qaEWFp9 pic.twitter.com/2Zhj6MH2dj
— PrakashLab (@PrakashLab) July 3, 2020
They were motivated to find diagnostics that tapped into new supply chains for testing. A standard lab centrifuge was used, but Cepko and Rabe invited others to improve the protocol for this step.
The Stanford lab’s Handyfuge is based on a flashlight toy with a unidirectional ratchet inside that Prakash discovered his children had. The Handyfuge follows a paper centrifuge that his group developed in 2017.
Centrifugation is important for separating components in biological samples. Collected saliva from a person suspected to have Covid-19 is first chemically inactivated. The tube is then incubated in boiling water for five minutes, to denature viral membranes and proteins.
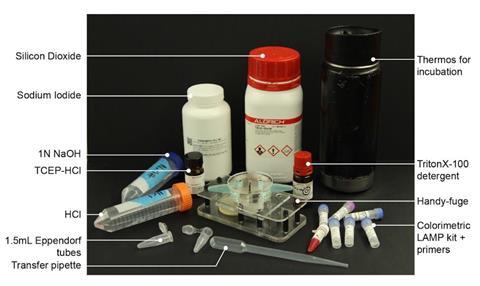
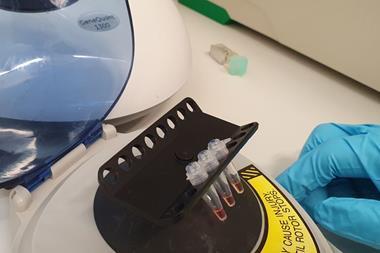
The inactivated sample is placed in the Handyfuge and spun at max speed for one minute by pumping the handle. A precipitate collects at the bottom of the tube, and the cleared supernatant containing inactivated virus is poured into a new tube. Salt binder is added and commercially available silica beads used to capture the nucleic acids from the saliva.
After more centrifuging and an ethanol wash, the Lamp primers and other reagents are added. A successful polymerisation reaction substantially reduces the pH of the solution, enabling detection of viral RNA as a change in colour from pink to yellow.
The Stanford lab is now talking to manufacturers to build the first 10,000 Handyfuges. The next big step, Prakash says, is to validate the test in the field. ‘Once that is done, we will apply for regulatory approval.’
Most Sars-CoV-2 tests rely on quantitative PCR assays, requiring multiple steps and precise temperature control, with robotics and expensive machines. The Lamp protocol with Handyfuge, however, is suitable for low resource settings which may not require high throughput. Lamp can detect viral RNA at less than 100 copies per microlitre. The reaction takes half an hour and gives unambiguous results.
Cepko was delighted to see the Handyfuge. ‘There are lots of motivated clever people working on these problems and solutions are being found,’ she says. ‘Now it is a matter of logistics and politics to get them where we need them.’ Anyone with technician training or a bachelor’s degree who can handle samples and pipette could run the assay, she adds.
The low-cost centrifuge is part of Prakash’s ‘frugal science’ effort to develop inexpensive tools, such as his widely available $1 Foldscope microscope. It is a platform technology and will also be used for future inexpensive diagnostics, such as for malaria or food adulteration.
References
1 E Li et al, medRxiv, 2020, DOI: 10.1101/2020.06.30.20143255
2 B A Rabe and C Cepko, medRxiv, 2020, DOI: 10.1101/2020.04.23.20076877















No comments yet