Biosensor runs off cells’ chemical fuel
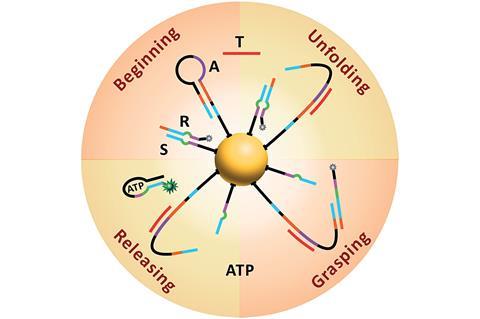
Scientists have designed a nanomachine that can enter a cell and, using endogenous ATP, move and fluoresce in the presence of a specific miRNA sequence.
Nanomachines have attracted wide attention for their potential in imaging, computing and other applications within living cells. Natural protein motors exist to aid muscle contraction and cellular transport but scientists have been unable to design efficient synthetic replicates.
One of the major challenges is delivering a machine and its fuel source into a cell and controlling their distribution. Now, Bin-Cheng Yin and colleagues at the East China University of Science and Technology, China, have for the first time designed a gold-nanoparticle based machine that moves using cells’ own power source, ATP.
Wenrong Yang, an expert in nanoscale biosensors at Deakin University, Australia, says ‘this study reports a smart and highly integrated DNA nanomachine that can be powered by endogenous ATP and operated inside living cells without any additives. These results open up a new avenue for the design and fabrication of new simple and effective DNA nanomachines’.
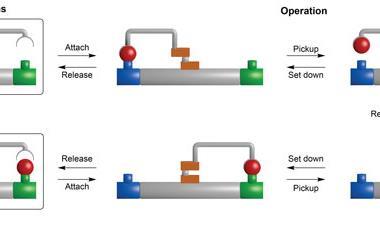
Yin’s group immobilised two different components onto a gold-nanoparticle; a hairpin loop (A) that contains the complementary sequence of the miRNA target, and hundreds of two-stranded DNA substrate complexes (S/R) that are densely packed and form a track. In the presence of the target sequence, the structure of A unloops into a freely swinging arm that attaches to R and breaks the S/R strand connection. ATP then fuels the release of the R strand leaving the swinging arm open to attach to more S/R strands. This motion moves the nanomachine along the track and creates a detectable fluorescence providing highly sensitive imaging of the target miRNA.
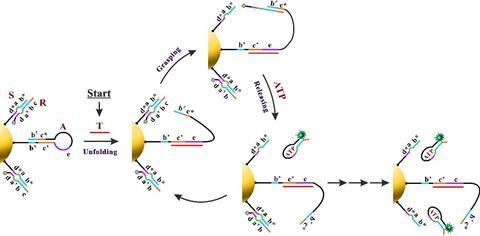
‘Importantly, the design principle of our ATP-powered DNA nanomachine is highly generalisable and can be easily adapted to execute complex tasks in living cells by customizing the machine components in response to different intracellular stimuli,’ says Yin. ‘For example, by simply changing the recognition sequence on the swing arm (A) into other miRNA antisense sequences or functional nucleic acids (such as aptamers), various targets such as miRNA, proteins and small molecules can be imaged.’
The team are now looking at applying their nanomachine to image clinically relevant molecules and act as a drug delivery vehicle for controlled therapy.













No comments yet