Non-natural system uses covalent base-pairing to transfer information
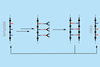
Template-directed process that replicates non-biological molecules is step towards using evolution to explore chemical space
Scientists have developed a purely synthetic system for replicating information from one generation to the next. It copies information encoded in monomer building blocks from a parent, template sequence to a daughter sequence and is the first transfer of sequential information in a system that doesn’t interface with DNA.