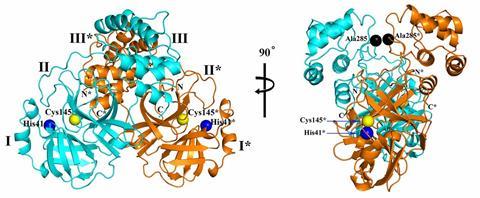
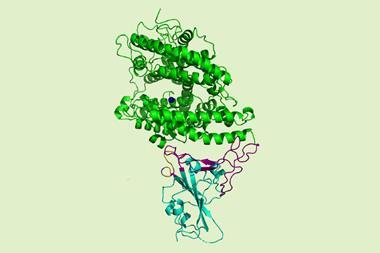
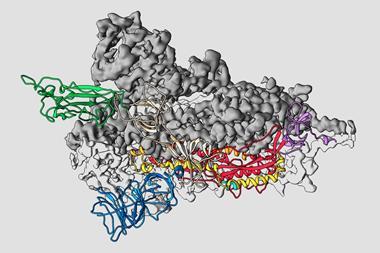
The three-dimensional architecture of the main protease of Sars-CoV-2 has been solved for the first time.1 The structural details open up a path to a therapy for Covid-19 by creating inhibitors for this enzyme that would impair viral replication.
The World Health Organization declared the outbreak of Covid-19 a pandemic on 11 March. As of 27 March, there were over 530,000 confirmed cases and 24,075 deaths.
The main Sars-CoV-2 protease is an enzyme that is crucial for the virus. It processes a large polyprotein – around 790kDa – that the virus makes by hijacking our own cell’s machinery, and then cleaves it into 12 smaller proteins that include the machinery the virus needs to replicate itself. The team at the University of Lubeck previously developed broad-spectrum inhibitors against this protease, known as α-ketoamides, for the major two types of coronaviruses that infect humans.2
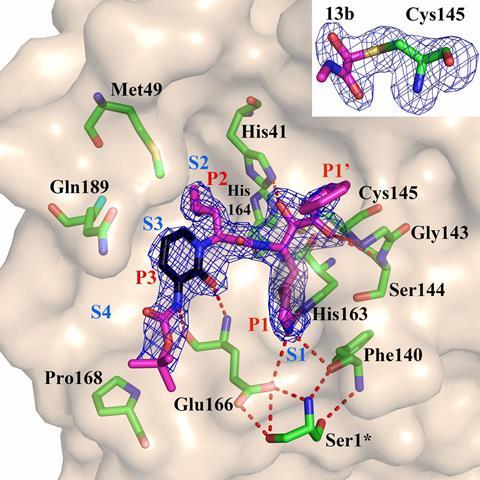
To better tailor these inhibitors to Sars-CoV-2, the team led by virologist Rolf Hilgenfeld used the high-intensity x-ray light source from the Berlin synchrotron source Bessy II to elucidate the 3D structure of the main protease. The crystal structures, including complexes between the protease and α-ketoamide inhibitors, offer a starting point for the development of new inhibitors or drugs that target the enzyme. These drugs could hit specific points on the protein and impede its function, inactivating the virus or at least making it far less lethal.
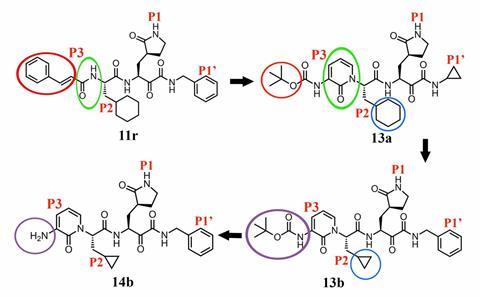
One compound, α-ketoamide 13b, inhibited the main protease of the virus, as well as the related Mers virus. The 13b inhibitor developed in Hilgenfeld’s lab was given to mice using a nebuliser without any adverse effects. This suggests direct delivery to the lungs may be feasible in Covid-19 patients.
In order to get the compound to persist for longer in the body, Hilgenfeld’s team’s original inhibitor was modified to hide an amide bond within a pyridone ring. This turns the compound from a substrate for the enzyme into an inhibitor, and also protects it from destruction by human proteases. The hydrophobicity of the compound was also tweaked, to reduce its binding to plasma proteins and boost solubility.
‘The virus’s major protease has been shown to be essential in other related viruses and as there are no identical proteases in the human body, it is a viable target,’ notes biochemist Charles Craik at the University of California San Francisco. Perhaps the best example of protease inhibitors are those against HIV.
‘This is an excellent starting point,’ adds Craik, referring to the 13b inhibitor. ‘More drug-like properties have to be adopted to avoid metabolism of the drug, improve its ability to cross membranes and get to the viral enzyme inside an infected cell.’
Increased specificity for the Sars-CoV-2 protein would also be beneficial. There are no such proteases in people, but ‘there are closely related enzymes that carry out important functions. Inhibiting them could cause serious side-effects.’
References
1 L Zhang et al, Science, 2020, DOI: 10.1126/science.abb3405
2 L Zhang et al, J. Med. Chem. 2020, DOI: 10.1021/acs.jmedchem.9b01828












No comments yet