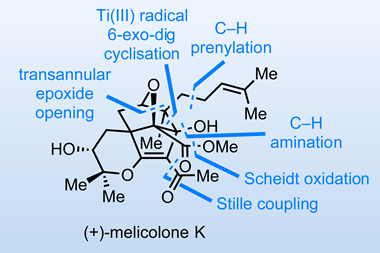
In the two decades since the Human Genome Project was declared finished (if not complete), DNA sequencing technology has advanced to the point where reading whole genomes is becoming routine. In 2003 the cost of sequencing an individual’s complete genome was in the order of millions of dollars, today it’s a few hundred. And that has led to huge growth in the size of genome databases.
What we can do with that data – what our genes tell us about our wellbeing and health, or how we behave and what we look like – is still in its infancy, however. In this issue, our cover feature looks at one of the ways the predictive potential of genomic data is being applied to catch criminals via forensic DNA phenotyping (FDP). It could be a powerful tool, and it also raises familiar ethical and societal questions about the benefits and harms of mining our genes.
As our recent feature explains, FDP goes beyond the well-established function of DNA forensics in confirming the identity of a suspect and evidence of their presence at a crime scene. Now, police forces have the capability to derive phenotypic fragments – hair colour, age, ethnicity – from a DNA sample. This tool can’t yet be used in court, but has been used by police forces to narrow down pools of suspects when matching a sample against DNA databases has come up empty handed.
The technique is powered by genome-wide association studies – large-scale statistical analyses of genomes that look for links between genes and disease risk, or behavioural or physiological traits. These studies are becoming faster and more reliable as they have greater volumes of data to work with, but they still tell us little about what goes on between the gene and its ultimate expression in a trait. What the studies do show is that these relationships are much more complicated than we had assumed. Large numbers of genes can be associated with even seemingly simple traits, and even in the very rare case where a gene is responsible for a single outcome, it’s only ever as a probability – they can’t begin to account for everything else that happens as a genome is expressed as a living person.
Yet at a shallower interpretation, these studies can seem to endorse the idea of ‘genes for’ a particular trait – a notion with concerning implications. Studies have shown that intelligence is a strongly heritable trait, for example, but how should we use that information as a society? Should parents be allowed to select or even edit embryos to yield the ‘best’ genome? There are also other issues of fairness and inclusion to address. The genome databases that exist today are heavily biased towards individuals of European descent, for example, so diversifying that pool is essential to allow everyone to benefit from such studies. Earlier this year the pharma companies AbbVie, Amgen, AstraZeneca, Bayer and Merck joined the Alliance for Genomic Discovery to fund genome sequencing of under-represented populations.
The barriers to widening participation in these programmes aren’t merely logistical, however. As the experts in our feature caution, technologies like FDP come with ethical concerns such as the risk of profiling and discrimination. In the US, genetic discrimination by employers or insurers is prohibited by law and many other countries have similar legislation, but it is far from being universal. The Australian insurance industry only recently adopted a moratorium on the use of genetic information in life insurance underwriting, for example. Data security poses a similar problem – in the EU, storing genetic information is covered by GDPR legislation, so it must be anonymised, kept private and secure, and individuals must typically consent to its use. Legal protections in other jurisdictions vary widely.
Legitimate concerns over how genomic information might be used dissuades individuals from participating in these studies. But the power of genomic data comes from the size and diversity of the dataset – so if people are to participate then they need to do so with informed consent and the surety of stringent safeguards.











No comments yet