Artificial intelligence tool to help doctors prioritise treatment
A team of researchers in the US and China has devised a severity scoring system for Covid-19 by looking at the biomarkers present in blood. The system combines biomarker measurements with risk factors such as age and gender in a statistical learning algorithm to predict the likelihood of a Covid-19 patient dying from the disease. It is the first quantitative point-of-care diagnostic tool to predict the severity of the disease in individual patients.
The system can detect several biomarkers from a single blood sample without needing to be sent away to a laboratory. Each test runs in a single-use microfluidic cartridge placed within a biosensor platform, which generates immunofluorescent signals that correlate to antigen concentrations.
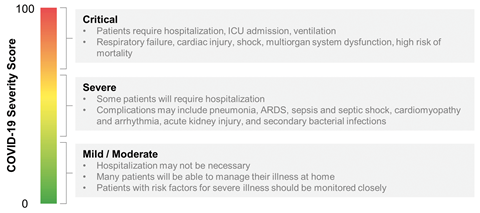
The biomarkers were specifically chosen because their presence was linked to poor outcomes in patients. Biomolecules such as D-dimer (a breakdown product of blood clots), C-reactive protein (a marker of inflammation or infection) and procalcitonin (increased levels of which indicate a bacterial co-infection and sepsis) were identified as relevant to complications associated with Covid-19, and were dramatically increased in patients that died versus those that recovered. The result is a score between 0–100 – a single number output is important, as a doctor can quickly identify whether a specific patient may be more or less likely to suffer life-threatening complications.
The use of severity score systems to make clinical decisions is well known – the team had previously worked on a severity score system for cardiovascular events, and they were able to use this body of research to help them when selecting biomarkers for Covid-19. John McDevitt, who led the project at New York University, was stunned that ‘a good scorecard did not exist’ for Covid-19. The researchers recognised that doctors did not have a good way to prioritise resources, and wanted to change that.
The algorithm was trained using an artificial intelligence approach and known outcomes from hospitalised Covid-19 patients in Wuhan. The model continues to learn – in fact, the slowest step in its development is acquiring the data. McDevitt says that once they are able to access data, new understanding comes ‘within days.’ Currently they are working with communities in Brooklyn and seeing how much influence cultural and economic differences have on the severity of the disease.
Brian Cunningham, an expert in biosensors and engineering at the University of Illinois at Urbana-Champaign, US, notes that the model can ‘never be fully predictive, even when it is used to evaluate a group of patients with similar characteristics.’ However, he believes that this tool could help identify high risk patients, allowing them to access further care ‘which could potentially save their life’.
The next steps are ambitious – the team are working on a free app for immediate release to help clinicians manage their patients, and they are adamant they don’t want this technology to only be available to the world’s richest medical centres.
Utkan Demirci, who specialises in applying nanoscale technologies to problems in medicine at Stanford University, US, says technologies similar to this one ‘will play a significant role [in future] as we combine these tools with existing health data and history at a personalised level merged with machine learning methods.’
References
This article is open access
M P McRae et al, Lab Chip, 2020, 20, 2075 (DOI: 10.1039/d0lc00373e)










No comments yet