Artificial nanopore proteins that resemble the natural ion channels found in cell membranes have been synthesised by researchers in the US and Europe. The team has shown how these nanopores can be built into sensors that detect medically relevant substances.
Ions cannot naturally pass through lipids. The effective functioning of cells, however, requires them to pass through cell membranes, which is where pore-forming proteins called ion channels come into play. ‘Imagine creating a water-filled space inside the lipid that the ion can go through like they would go through normal solution,’ says David Baker, the director of the Institute for Protein Design at the University of Washington in Seattle, US.
These features can potentially be useful for detecting substances like toxins, and researchers have therefore tried to modify the shapes and sizes of natural ion channels. However, the flexibility of this approach is limited. One problem is that deep learning algorithms like Deepmind’s AlphaFold and the Institute for Protein Design’s own RoseTTAFold have relatively little training data. ‘About 30% of the proteins in our body fall in membranes, but it’s very difficult to work with membrane proteins in the lab because they need to be solubilised in lipids,’ explains Anastassia Vorobieva at Vrije Universiteit Brussel in Belgium. ‘So we don’t have an awful lot of knowledge about how these proteins actually fold and function.’
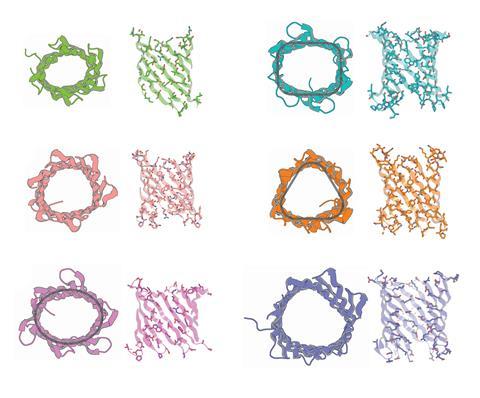
Now, a team led by Baker and Vorobieva has used a biophysics-based approach to design pore-forming proteins from scratch using interconnected polypeptide chains called β-strands.1 The researchers had produced eight-stranded ‘β-barrels’ in 2021, but these were too narrow to contain central conducting channels. The researchers reasoned that increasing the number of interwoven strands would widen the channels.
Hypothetically optimal designs, in which all the hydrophilic groups faced inwards, almost never formed the desired structures because proteins that aggregated too readily simply formed a clump. The team therefore incorporated some inward-facing hydrophobic groups to frustrate structure formation. ‘It’s very fun because we work with these tools in a way that’s very different from everyone else,’ says Vorobieva. ‘We really need to introduce these sub-optimal positions with tools that are just meant to find optimum positions.’
The researchers produced β-barrels comprising 10, 12 and 14 strands with progressively larger pores and also varied their shapes. They then measured the electrical conductivity across phospholipid membranes into which the β-barrels were incorporated. ‘You have electrodes on both sides of this membrane, and you throw in your protein,’ says the University of Washington’s Sagardip Majumder, who also worked on the project. ‘When there is a pore, the ions can pass through, and you get some current.’
Small molecule sensors
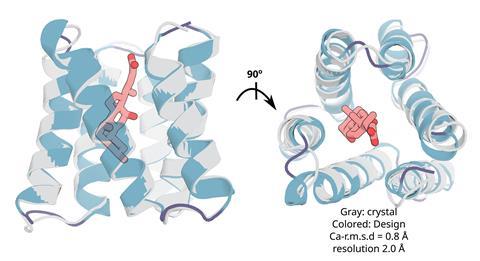
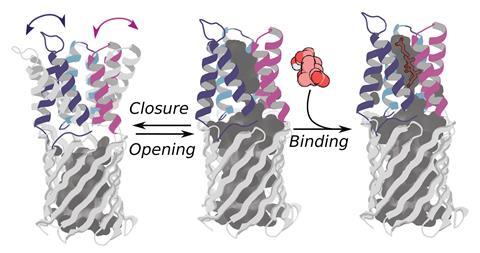
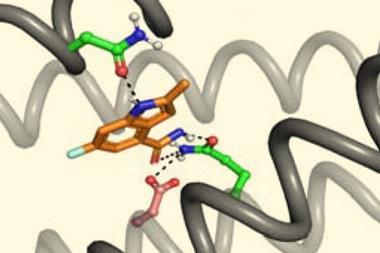
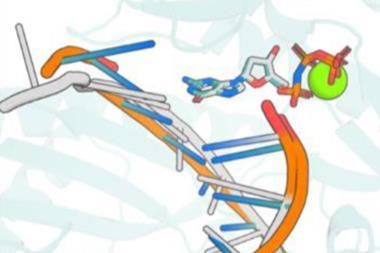
The researchers then showed how these nanopore proteins can be used in the detection of small molecules such as cholic acid, an indicator of liver disease, and the anticancer drug methotrexate.2 With the aid of their recently developed LigandMPNN deep learning-based algorithm, they designed ‘pseudocyclic’ proteins that bind specific small molecule ligands. They then divided the pseudocyclic proteins into fragments and attached these to their nanopore proteins.
In the presence of the target molecules, the small molecule ligands would bind all the fragments and stick the pseudocyclic proteins back together. These reassembled proteins would then temporarily block the β-barrel ion channels that they were connected to. By measuring changes in the conductivity of the ion channel, the researchers could detect the presence of a target molecule and its concentration.
The researchers hope the method could find various applications, such as home testing of drugs and toxins that currently require routine clinical monitoring. ‘You really don’t want the level [of methotrexate] to go above a certain value or else you’ll [die], so we created a binder for it in the hope of turning that into a sensor,’ says the University of Washington’s Linna An, who led the work on the sensor design.
Harvard Medical School’s Nicholas Polizzi, who wasn’t involved in the work, hails the design of the artificial nanopores as a ‘step change’. ‘The amount of protein biophysics that went in to understanding how to get these proteins to fold into a membrane … it seemed like a really cool puzzle that [Vorobieva’s team] has solved or at least worked out in large part.’
Polizzi notes that the hit rate for designing the ligand-binding pseudocyclic proteins was rather low, even with the researchers’ LigandMPNN algorithm. Nevertheless, he says it is ‘a really cool innovation to try and stick these on top of Anastassia’s nanopores to make them sensors for small molecules’ and describes the work as ‘a proof of principle’.
References
1. S Berhanu et al, Science, 2024, DOI: 10.1126/science.adn3796
2. L An et al, Science, 2024, DOI: 10.1126/science.adn3780












No comments yet