Lithium electrolyte helps aerolysin overcome electrostatic obstacle to detect ssDNA longer than 100 nucleotides
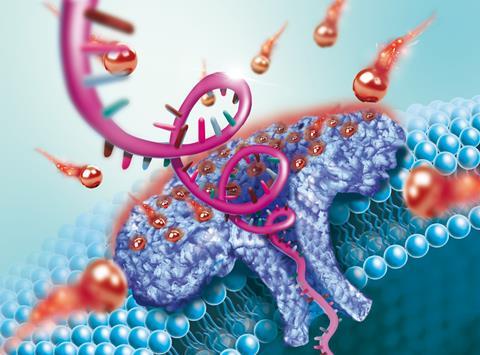
Scientists in China have used a bacterium-derived nanopore to effectively trap and detect single-stranded DNA over 100 nucleotides long for the first time.
‘Nature provides us with a most intelligent way to capture and sense individual molecules,’ explains Yi-Lun Ying of East China University of Science and Technology, Shanghai, who led the team behind the improved nanopore-based DNA sensing strategy. ‘This tiny interface can confine electrochemical effects, quantum effects, and magnetism, which may open up versatile possibilities for seeing the world from a single molecule view.’
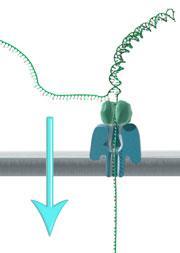
Industry already uses pore-forming toxins, such as staphylococcal hemolysin – which attacks blood cells, for sequencing small strands of DNA. The process works by passing an ionic current through the nanopore and identifies biological molecules translocating through the nanopore based on how those molecules alter the current.
Aerolysin, another bacterium-derived toxin, destroys eukaryotic cells. It also has an outstanding ability for blocking ionic currents when chemical structures translocate through its internal channel. Compared with hemolysin, aerolysin has several advantages, such as a higher current resolution and a more robust sensing system. In previous studies, however, DNA struggled to pass from one side of the aerolysin pore to the other.
‘Right now, the major obstacle for aerolysin is its short sensing length for DNA,’ explains Ying. Aerolysin captures ssDNA at a very low rate compared to other biological nanopores and can only detect very short oligonucleotides (<10 nucleotides). This limitation is mainly attributed aerolysin’s negatively-charged nanopore stopping the negatively-charged DNA from entering it.
Now, Ying and colleagues have overcome this obstacle by using a lithium electrolyte (LiCl), instead of the commonly used potassium electrolyte, to neutralise the surface of the nanopore. This allows DNA, especially long oligonucleotides (<102 nucleotides), to pass one-by-one through the pores in the aerolysin lipid bilayer. The result was an increase in the DNA capture rate by a factor of 10, whereupon the researchers identified the single molecule characteristics of each molecule from the resulting current blockade.
Fan Xia, who studies biomolecular responsive nanopores at the China University of Geosciences in Wuhan, says that the ability to capture long DNA molecules, closer to the size of realistic DNA samples, will have real world applications. ‘The longer the detectable single strand, the simpler the preliminary treatment that needs to be done to the actual DNA sample.’
References
This article is open access
Z Hu, M Li, S Liu, Y Ying and Y Long, Chem. Sci., 2019, 10, 354 (DOI: 10.1039/c8sc03927e)









No comments yet